Computational Immunology
In collaborations with investigators in the immunology and rheumatology fields, we focus on better understanding complex disease through single cell and spatial technologies. We have several projects harnessing multi-omic immunological profiling to characterize immune activation and heterogeneity in disease. Integrative analysis of data such as multi-parameter flow cytometry, cytokine production assays, single-cell gene expression of immune cell compartments and microbiome assays have powered deep phenotyping of complex immune processes.
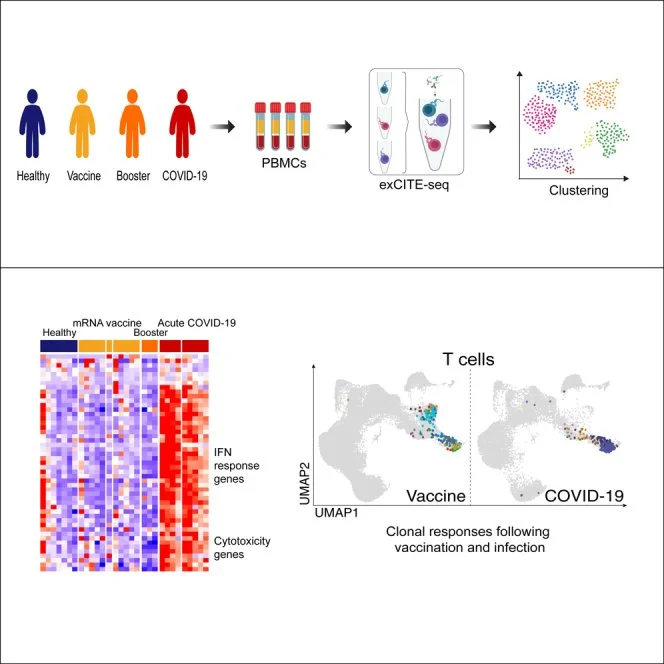
Ivanova et al., 2023
Recent Publications:
Ivanova EN et al., mRNA COVID-19 vaccine elicits potent adaptive immune response without the acute inflammation of SARS-CoV-2 infection (2023) iScience
Cornwell MG et al., Modeling of clinical phenotypes in systemic lupus erythmatosis based on the transcriptome and FCGR2a genotype. (2023) Journal of Translational Medicine
Zwack EE et al, Staphyloccocus aureus induces a muted host response in human blood that blunts the recruitment of neutrophils. (2022) PNAS
Devlin JC et al., Distinct Features of Human Myeloid Cell Cytokine Response Profiles Identify Neutrophil Activation by Cytokines as a Prognostic Feature during Tuberculosis and Cancer. (2020) Journal of Immunology
Cardiovascular Multiomics
As a collaborator of the ISCHEMIA trial, we are in the process of developing novel methods to analyze multi-omic data in an effort to characterize ischemic heart disease. Our primary goals and to draw significant causative conclusions that can further our understanding of the genomic and transcriptomic underpinnings of the clinical manifestations and development of ischemic heart disease. We are also involved in multiple studies to understand the role of platelet activity in disease pathology in collaboration with the Berger lab.
Recent Publications:
Newman JD et al., Biomarkers and cardiovascular events in patients with stable coronary disease in the ISCHEMIA Trials (2023) American Heart Journal
Barrett TJ et al., Platelets amplify endotheliopathy in COVID-19. (2022) Science Advances
Newman JD*, Cornwell MG* et al., Gene Expression Signature in Patients with Symptomatic Peripheral Artery Disease. (2021) Arteriosclerosis, Thrombosis, and Vascular Biology
Cancer Proteogenomics
We are focusing on the application of computational and statistical methods to better understand the mechanisms underlying tumor biology and metastasis. To do this, we use a number of high-dimensional data types including bulk, single cell and spatial technologies to analyze and integrate proteomics, genomics and transcriptomics of tumors to allow for better understanding of the static and dynamic status of the disease.
Dou and Kawaler et al., 2020
Recent Publications:
Schraink et al., PhosphoDisco: A Toolkit for Co-regulated Phosphorylation Module Discovery in Phosphoproteomic Data (2023) Molecular and Cellular Proteomics
Kleffman K. et al., Melanoma-Secreted Amyloid Beta Suppresses Neuroinflammation and Promotes Brain Metastasis (2022) Cancer Discovery
Krug K et al., Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy (2020) Cell
Gillette M.A. et al., Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. (2020) Cell